Format conversion using a galaxy tool
Initial Format (EMBL flat file)
ID INE1 standard; DNA; INV; 611 BP.
XX
AC U66884;
XX
DR FLYBASE; FBte0000312; Dmel\INE-1.
XX
SY synonym: mini-me
SY synonym: DINE
SY synonym: narep1
SY synonym: Dr. D
XX
FT source U66884:4880..15490
XX
CC This is presumably a dead element.
CC Derived from U66884 (e1371475) (Rel. 52, Last updated, Version 6).
CC Michael Ashburner, 28-Sep-2001.
CC Any changes to original sequence record are annotated in an FT line.
XX
SQ Sequence 611 BP; 193 A; 123 C; 93 G; 202 T; 0 other;
TATACCCGTT ACTAGATTCG TTGAAATGAA TGTAACAGGC AGAAGGAAGC GTCTTAGACC 60
ATATATAGTA TATACATACA TGTATATTCT TGATCAGGAT CAATAGCCGA GTCGATCTTG 120
CCATATCCGT CTGTCCGTAT GAACGTCGAG ATCTCAGGAA CTATAAAAGC TAGAAGGTTT 180
AGATTCAGCA TACAGAGACA AAGACGCAAG TAGCCATGCC CACTCTAACG TCCACAAACA 240
GCGCAAAACT ATCACGCCCA CACTTTTGAA AAATGTGTTG TTCTTTTCAC ATTCTGATTA 300
GTCTTTTACA TTTCTATCGA TTTCCAAAAA AAAACTTTTT GCCAACGCCC TAAAACCGCC 360
CAAAACTCCG ACACCCACAT TTGTAAAAAA TTGTTGGGAA TTTTTTTCAT AAATTTATTA 420
GTTTATTATT TATTATAAAT TTAAGTTTAT ATCGATTTGC CGACAACATA TTTTAATTTT 480
TTTTCTCATT TTATCTTTTA TCTATCGATA TCCCAGAAAA ATTGTGCAAT TTCGCATTCA 540
CACTAGCTGA GTAACGGGTA TCTGATAGTC GGGAAACTCG ACTATAGCAT TCTCTCTTTT 600
TGAAATTGCG G 611
//
Target Format (fasta)
>INE1
TATACCCGTTACTAGATTCGTTGAAATGAATGTAACAGGCAGAAGGAAGCGTCTTAGACC
ATATATAGTATATACATACATGTATATTCTTGATCAGGATCAATAGCCGAGTCGATCTTG
CCATATCCGTCTGTCCGTATGAACGTCGAGATCTCAGGAACTATAAAAGCTAGAAGGTTT
AGATTCAGCATACAGAGACAAAGACGCAAGTAGCCATGCCCACTCTAACGTCCACAAACA
GCGCAAAACTATCACGCCCACACTTTTGAAAAATGTGTTGTTCTTTTCACATTCTGATTA
GTCTTTTACATTTCTATCGATTTCCAAAAAAAAACTTTTTGCCAACGCCCTAAAACCGCC
CAAAACTCCGACACCCACATTTGTAAAAAATTGTTGGGAATTTTTTTCATAAATTTATTA
GTTTATTATTTATTATAAATTTAAGTTTATATCGATTTGCCGACAACATATTTTAATTTT
TTTTCTCATTTTATCTTTTATCTATCGATATCCCAGAAAAATTGTGCAATTTCGCATTCA
CACTAGCTGAGTAACGGGTATCTGATAGTCGGGAAACTCGACTATAGCATTCTCTCTTTT
TGAAATTGCGG
Import the dataset¶
- In galaxy, create a new history and name it "EMBL to Fasta conversion"
- Copy the url of the flat EMBL file:
- In Galaxy, click the
Upload Databutton
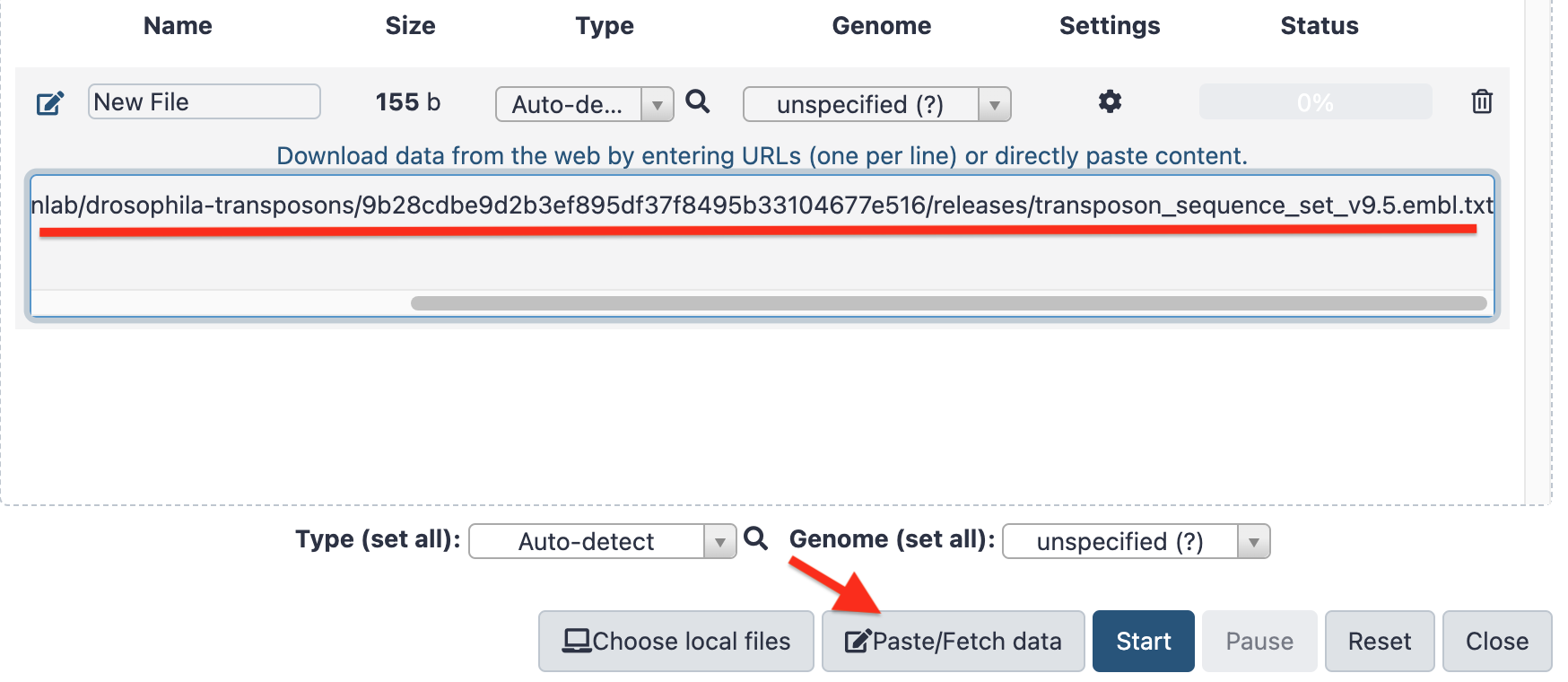
- Then click the
Paste/Fetch databutton, Paste the copied file url in the central field and clickStart
Reformat the file using the tool embl2fa:¶
- Go to the
Admin→Install and Uninstallpanel. - In the search repository box, type
embl2fa - The search should likely return the tool at the bottom of the page
- Click on the embl2fa, then on the
installbutton. - Choose
AG 2023for the Target Section:, then click theOKbutton - The tool installation should only take a few seconds (the button
Installturns to a redUninstall) - You can now go back to the analysis interface by clicking the
homeicon. - in the Galaxy search toolbar box, search for
embland select the toolConvert embl flat file to fasta. - Select the imported dataset
transposon_sequence_set_v9.5.embl.txt(should likely be the dataset #1) and clickRun Tool
Inspect the new dataset.¶
Inspect the new fasta file dataset by clicking the small rounded i icon that shows up
when you deploy the dataset.
In particular, you can deploy the Command line box in the datasheet and verify the code
executed by the tool.
Check the conversion¶
- Download the file reference for the conversion (ie, a file that we know is correctly converted...)
- Use the tool
Differences between two filesto compare the datasetfasta fileand the datasettransposon_sequence_set_v9.5.fa
The resulting dataset should be empty, meaning that the dataset fasta file and the dataset
transposon_sequence_set_v9.5.fa are identical.