Running a workflow in Galaxy
In this use case, we are going to
- Upload 2 workflow description files in the Galaxy server instance
- Visualise these workflows and see that tools to execute the workflows are missing
- since you are administrating the instance, install the missing tools
- Eventually run the workflows on input data obtained from a remote public repository.
Upload workflow description file (.ga)
- Ensure you are connected to your Galaxy server as an admin (the email you have entered in the galaxy.yml configuration file and the password to entered for this login when you registered for the first time)
- Click the workflow menu
- Click the "Upload or import workflow" button at the top right
- In the
Galaxy workflow URL:field, paste the url of the workflow file:
https://raw.githubusercontent.com/ARTbio/Run-Galaxy/master/workflows/Galaxy-Workflow-canonical_transposons.gtf_from_transposon_sequence_set.txt.ga
Note that this file is in the Run-Galaxy repository where all the material for this training is hosted
-
Click on the
Importbutton -
repeat the same operation with the other workflow
https://raw.githubusercontent.com/ARTbio/Run-Galaxy/master/workflows/Galaxy-Workflow-Extract_canonical_transposons_fasta.ga
(alternatively, you could upload the workflow files from you computer instead of uploading them by URL)
- Now, click on the workflows that are showing up in the workflow list:
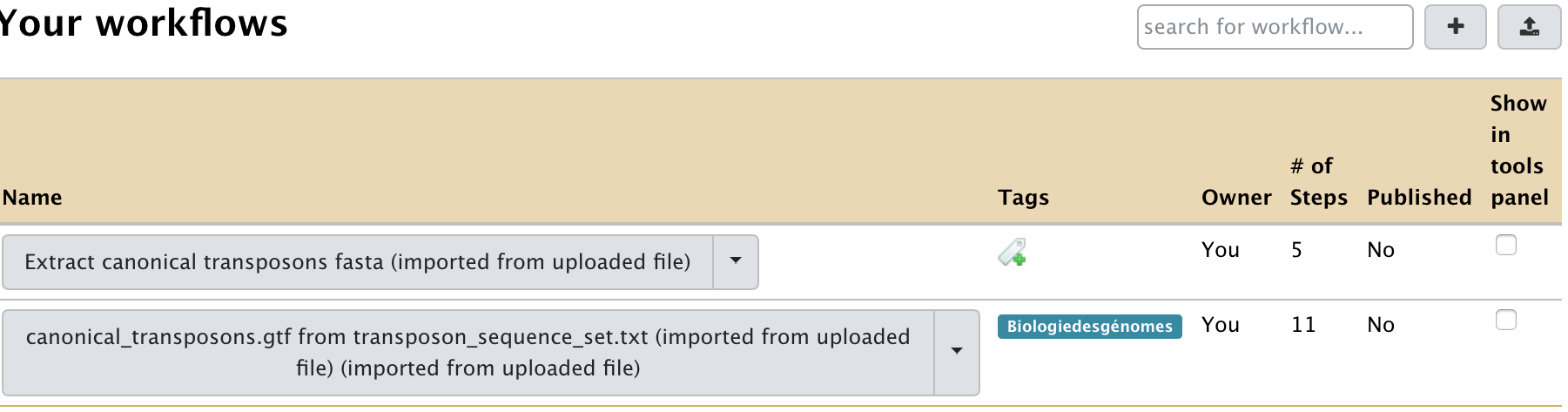
- Click a workflow and select the
Editoption - Observe the warning window that should look like:
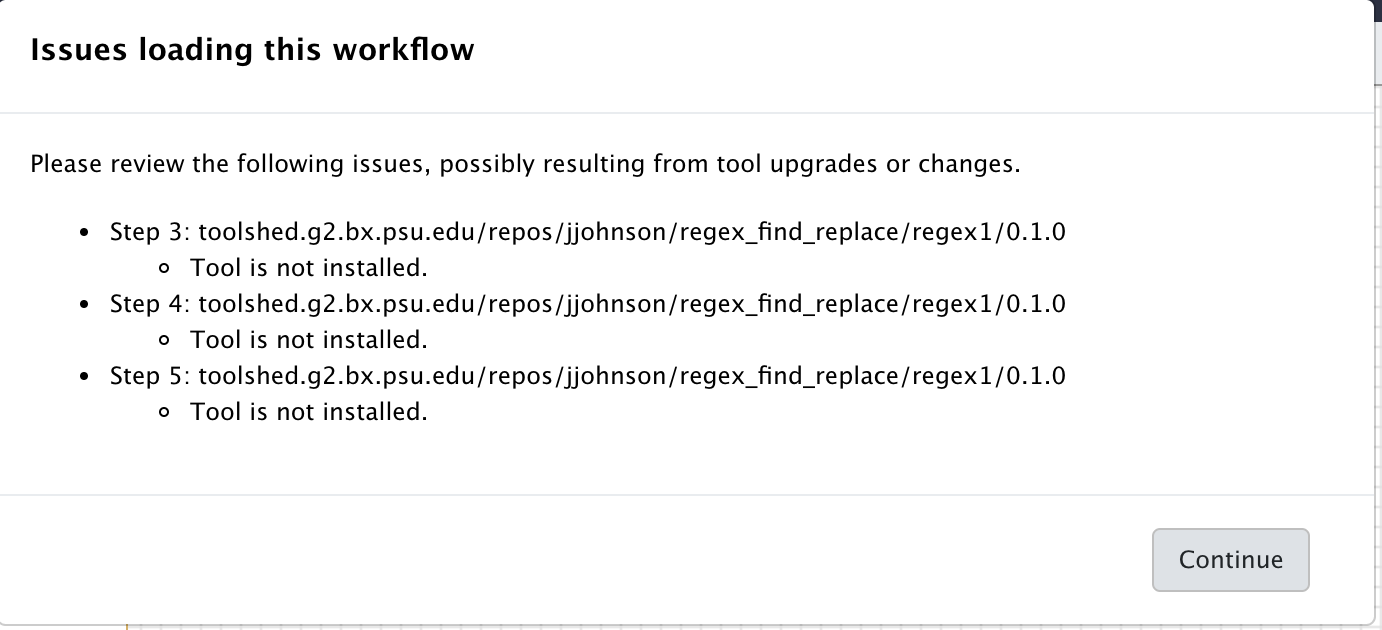
When you read the warnings, you will see that the workflow was indeed successfully imported. However, tools are missing, namely:
toolshed.g2.bx.psu.edu/repos/kellrott/regex_replace/regex_replace/1.0.0, version 1.0.0
toolshed.g2.bx.psu.edu/repos/blankenberg/column_regex_substitution/column_regex_substitution/0.1.0, version 0.1.0
toolshed.g2.bx.psu.edu/repos/jjohnson/regex_find_replace/regex_find_replace/0.1.0, version 0.1.0
The other lines are redundant, because the workflow is using the same tools at different steps.
Click on the Continue button. You should now see missing tools in red and missing links
between various workflow steps. Note that some tools are indeed present because they are
installed by default in the provided Galaxy framework.
-
We are going to fix this. Click on the
Continuebutton and then the upper "wheel" icon and selectClose, we will come back to the workflow editor when the missing tools are installed in the server. -
So far, so good, the missing tools are reported in the tools.yml file in the Run-Galaxy repository (or just above in a more complex format)
Installing missing tools
Thus, we have to install our first three tools in our Galaxy instance:
tools: - name: regex_replace owner: kellrott revisions: - 9a77d5fca67c tool_panel_section_label: biologie des genomes tool_shed_url: https://toolshed.g2.bx.psu.edu
- name: column_regex_substitution owner: blankenberg revisions:
-
12b740c4cbc1 tool_panel_section_label: biologie des genomes tool_shed_url: https://toolshed.g2.bx.psu.edu
-
name: regex_find_replace owner: jjohnson revisions:
-
9ea374bb0350 tool_panel_section_label: biologie des genomes tool_shed_url: https://toolshed.g2.bx.psu.edu
-
Click on the
Admintop menu - On the left bar click on
Manage tools
Check that there is actually no installed tools !
- Now, click the
Search Tool Shedmenu (again in the left bar) - Press the
Galaxy Main Tool Shedbutton - In the search field, copy and paste
regex_replace, and press theenterkey. - One tool will show up, owned by
kellrott. Click this tool, and selectpreview and install(No other solution anyway) - Click the
Install to Galaxybutton at the top of the screen - In the
Select existing tool panel section:menu, selectText Manipulation. Thus, the tools will appears in the sectionText Manipulationof the Galaxy tools. - Click
Install - You are going to wait for ~20 sec or so, before seeing the
Monitor installing tools...screen. - Rapidly enough, the Installation status should turn out green.
-
Click again the
Manage toolsmenu in the left bar, and look at the newly installed toolregex_find_replacein the list. -
Repeat the same operations for the tool
regex_find_replaceowned byjjohnson(version1.1.0). Do not take the tool with the same name but owned bygalaxyp -
Repeat the same operations for the tool
column_regex_substitutionowned byblankenberg(version0.0.1)
For this last installation, you will see a different panel after clicking Install to Galaxy:
If you scroll down a little bit, you should see a list of uninstalled tool dependencies like this:
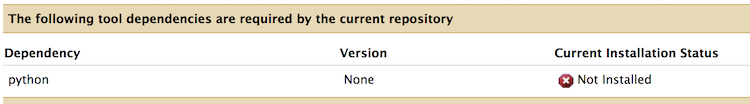
This is a software package required to get the tool column_regex_substitution working properly.
The required package (python 2.7) will be installed by the package manager conda.
You can further check this by clicking the Display Details button bellow the Dependency list.
At this stage, avoid further distraction and do not forget to select tool panel section
Text Manipulation, and finally click the Install button.
This time, the Monitor installing tool shed repositories will display new steps (in yellow),
including the Installing tool dependencies step. The whole process may take longer,
but not too long in this specific case.
- Finally go back a last time to the Manage tools panel:

There you see all three tools needed to properly run the imported workflow.
Check that the imported workflows now display correctly
If you click the workflow top menu, you should now be able to edit the imported workflows,
and see that everything is displaying correctly. For the workflow
canonical_transposons.gtf from transposon_sequence_set.txt :
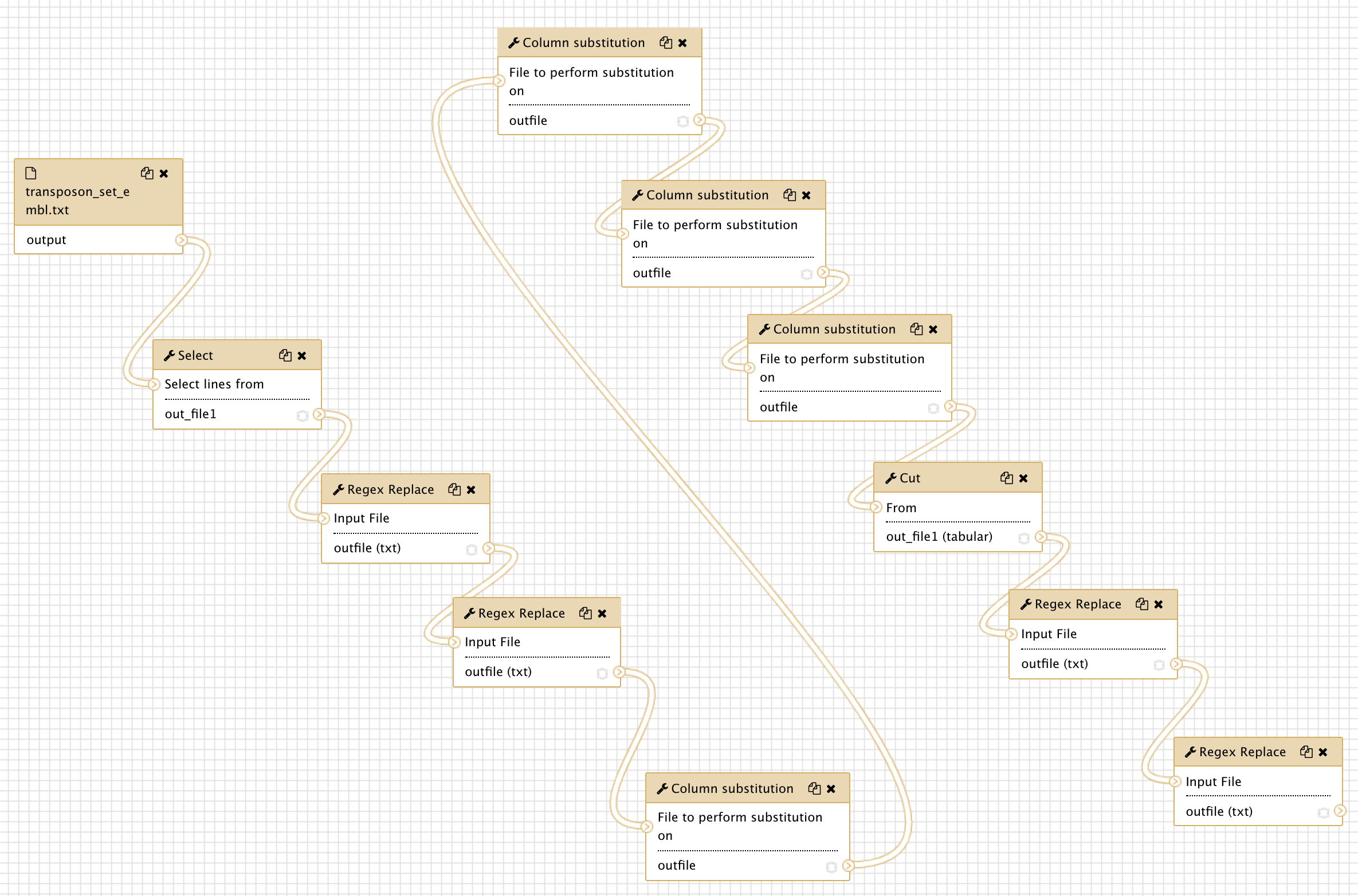
We can go through the various steps of the workflows and figure out what they are doing.
This first workflow performs a suite of find-and-replace text manipulations, starting
from input data that has been tagged transposon_set_embl.txt and producing a new text
dataset that is renamed canonical_transposons.gtf.
The second workflow uses the same input data file transposon_set_embl.txt to generate
a fasta file of canonical_transposon sequences
We will come back to all these steps after the workflows execution. However, we need to retrieve the input data set before running the workflow on these data.
Retrieve the transposon_set_embl.txt dataset
- Create a new history and name it
transposon_set_embl.txt manipulation - import the dataset using the
Paste/Fetch datamode of the upload manager (the small bottom-top arrow icone at the top left of the Galaxy interface). Copy the URLhttps://github.com/cbergman/transposons/raw/master/current/transposon_sequence_set.embl.txtin the open field and click theStartbutton. - have a close look at the file
Run the workflow
- Click on the workflow menu
- Click on the first workflow and select the Run option
- Leave the
Send results to a new historymenu to theNooption for the moment. - Just Click the
Run workflowbutton to run the workflow, and look at datasets in the history turning from grey to yellow to green. Note: often you don't see the dataset in the "yellow" state (running). You just need to refresh the history with the 2-curved-arrows icon of the local history menu. - repeat the same operation (from the input history) for the second workflow
Extract canonical transposons fasta (imported from uploaded file)