
HISAT2 (option for 50 % of attendees)

- create a new history and name it
HISAT2 - Import the 11 datasets from the RNAseq data library to this
HISAT2history, plus the Drosophila_melanogaster.BDGP6.95.gtf file - Select the
HISAT2tool with the following parameters to map your reads on the reference genome:Source for the reference genome: Use a builtin genomeSelect a reference genome: dm6Is this a single or paired library:Single-EndFASTA/Q file:GSM461176_untreat_single.fastq.gzGSM461179_treat_single.fastq.gzGSM461182_untreat_single.fastq.gz
Specify strand information:UnstrandedSummary optionsOutput alignment summary in a more machine-friendly style.: YESPrint alignment summary to a file.: YES
- Leave other settings as defaults
Execute

Redo the HISAT2 run for paired-end files
- Rerun the
HISAT2tool with the following parameters to map your reads on the reference genome:Source for the reference genome: Use a builtin genomeSelect a reference genome: dm6Is this a single or paired library:Paired-EndFASTA/Q file #1:GSM461177_1_untreat_paired.fastq.gzGSM461178_1_untreat_paired.fastq.gzGSM461180_1_treat_paired.fastq.gzGSM461181_1_treat_paired.fastq.gz
FASTA/Q file #2:GSM461177_2_untreat_paired.fastq.gzGSM461178_2_untreat_paired.fastq.gzGSM461180_2_treat_paired.fastq.gzGSM461181_2_treat_paired.fastq.gz
Specify strand information:Unstranded- Leave other settings as defaults (since you are redoing a run)
Execute
Rename your datasets !

You need now to rename you datasets to facilitate your downstream analysis.
Be quiet and focus ! No hurry, this is an important task in the analysis.
-
Search and select datasets with HISAT2
-
Click on the info icon
 of both
of both (BAM)andMapping summaryfiles -
Copy the name or one of the two names of the datasets as shown bellow

- Now click on the pencil icon of the same dataset

- Paste your text in the
Namefield of the dataset
- Edit your text as follow for
Mapping summaryfiles
-
Edit your text as follow for
(BAM)files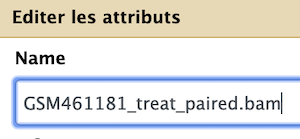
-
repeat ad lib for all Mapping summary and (BAM) files